GigaLab is for researchers changing the world in
AI m
Leveraging Parse’s innovative Evercode technology and expanded automation, GigaLab enables groundbreaking research never before deemed possible with current single cell approaches. With GigaLab, you’re enabled to explore greater than 10M cells in a single run. It’s real. And it’s real big.
Sequence 10M Cells Per Run In the GigaLab
The need for larger, more powerful studies is here. In the past 2 years, the average single cell count in publications is doubling each year, with the number of cells per study now well over the million cell mark. But how do we continue to expand the power of these studies?
Enter the Parse GigaLab – the world’s first and only solution to meet this demand head-on. With the unprecedented ability to profile over 10 million cells or nuclei in a single run, it’s redefining the boundaries of what’s possible in single cell research.
The need for bigger datasets is especially critical in areas like Drug Discovery research and screening, where the ability to visualize more drug/cell perturbations is incredibly empowering. This data is feeding large machine learning algorithms that will better help us to predict how individuals may respond to a given therapy, just based on the information in their cells.
Exponentially scalable
With the capacity to analyze 2.5B cells per year, GigaLab provides unlimited scalability for even the largest studies
Unmatched data quality
With robust Evercode™ technology as its core, GigaLab datasets include better detection of low expressors, and avoid common pitfalls of droplet-based single cell approaches
Exceptional Speed
With barcoding and library prep of 10M cells across up to 1,152 samples taking less than 3 days, GigaLab data is generated at a speed and scale never demonstrated before
Screening Cytokines Across 1000 Samples and 10M Cells in One Experiment
How we did it: Experimental design and method
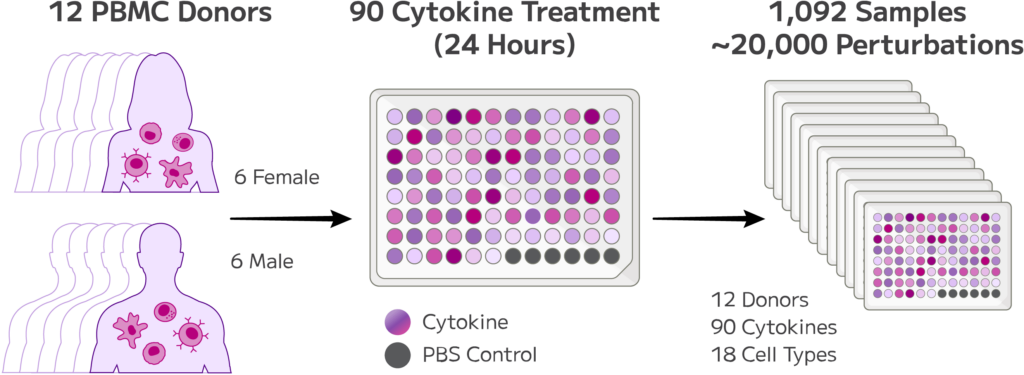
Starting with 12 donors, this first of its kind study measured 90 cytokine perturbations across 18 immune cell types, resulting in nearly 20,000 observed perturbations. This generated a 10 million cell dataset with 1,092 samples in a single run, with all barcoding and library preparation taking just 3 days. Once prepared, the libraries were sequenced on the Ultima UG100 instrument in partnership with the Ultima Genomics team.
More cells means more differentially expressed genes
Differential expression analysis was performed and the number of genes with a positive log fold change of > 0.3 and an adjusted p value of < 0.001 relative to the PBS control was calculated for every donor and cell type. Donors were ordered from left to right female to male and younger to older.
